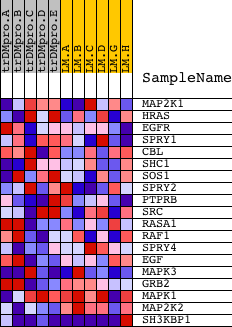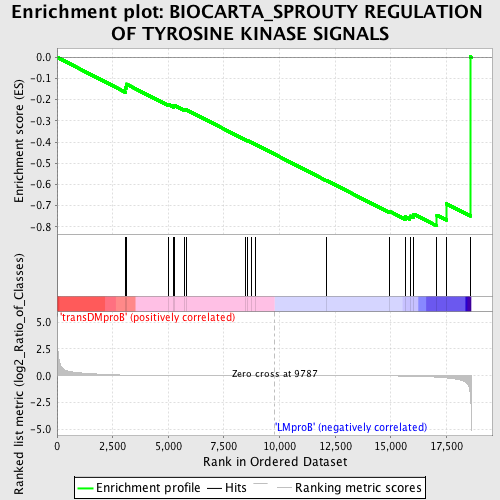
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | BIOCARTA_SPROUTY REGULATION OF TYROSINE KINASE SIGNALS |
| Enrichment Score (ES) | -0.79580176 |
| Normalized Enrichment Score (NES) | -1.473936 |
| Nominal p-value | 0.053452116 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MAP2K1 | 19082 | 3065 | 0.055 | -0.1441 | No | ||
| 2 | HRAS | 4868 | 3102 | 0.053 | -0.1260 | No | ||
| 3 | EGFR | 1329 20944 | 4992 | 0.015 | -0.2218 | No | ||
| 4 | SPRY1 | 15606 1901 | 5245 | 0.013 | -0.2303 | No | ||
| 5 | CBL | 19154 | 5255 | 0.013 | -0.2258 | No | ||
| 6 | SHC1 | 9813 9812 5430 | 5728 | 0.011 | -0.2471 | No | ||
| 7 | SOS1 | 5476 | 5800 | 0.010 | -0.2470 | No | ||
| 8 | SPRY2 | 21725 | 8458 | 0.003 | -0.3889 | No | ||
| 9 | PTPRB | 19875 | 8579 | 0.002 | -0.3945 | No | ||
| 10 | SRC | 5507 | 8725 | 0.002 | -0.4016 | No | ||
| 11 | RASA1 | 10174 | 8933 | 0.002 | -0.4121 | No | ||
| 12 | RAF1 | 17035 | 12094 | -0.005 | -0.5802 | No | ||
| 13 | SPRY4 | 23448 | 14922 | -0.018 | -0.7253 | No | ||
| 14 | EGF | 15169 | 15648 | -0.032 | -0.7521 | Yes | ||
| 15 | MAPK3 | 6458 11170 | 15861 | -0.039 | -0.7487 | Yes | ||
| 16 | GRB2 | 20149 | 16030 | -0.047 | -0.7400 | Yes | ||
| 17 | MAPK1 | 1642 11167 | 17069 | -0.135 | -0.7451 | Yes | ||
| 18 | MAP2K2 | 19933 | 17495 | -0.205 | -0.6909 | Yes | ||
| 19 | SH3KBP1 | 2631 12206 7189 | 18578 | -1.998 | 0.0020 | Yes |